How to use MolGpKa?
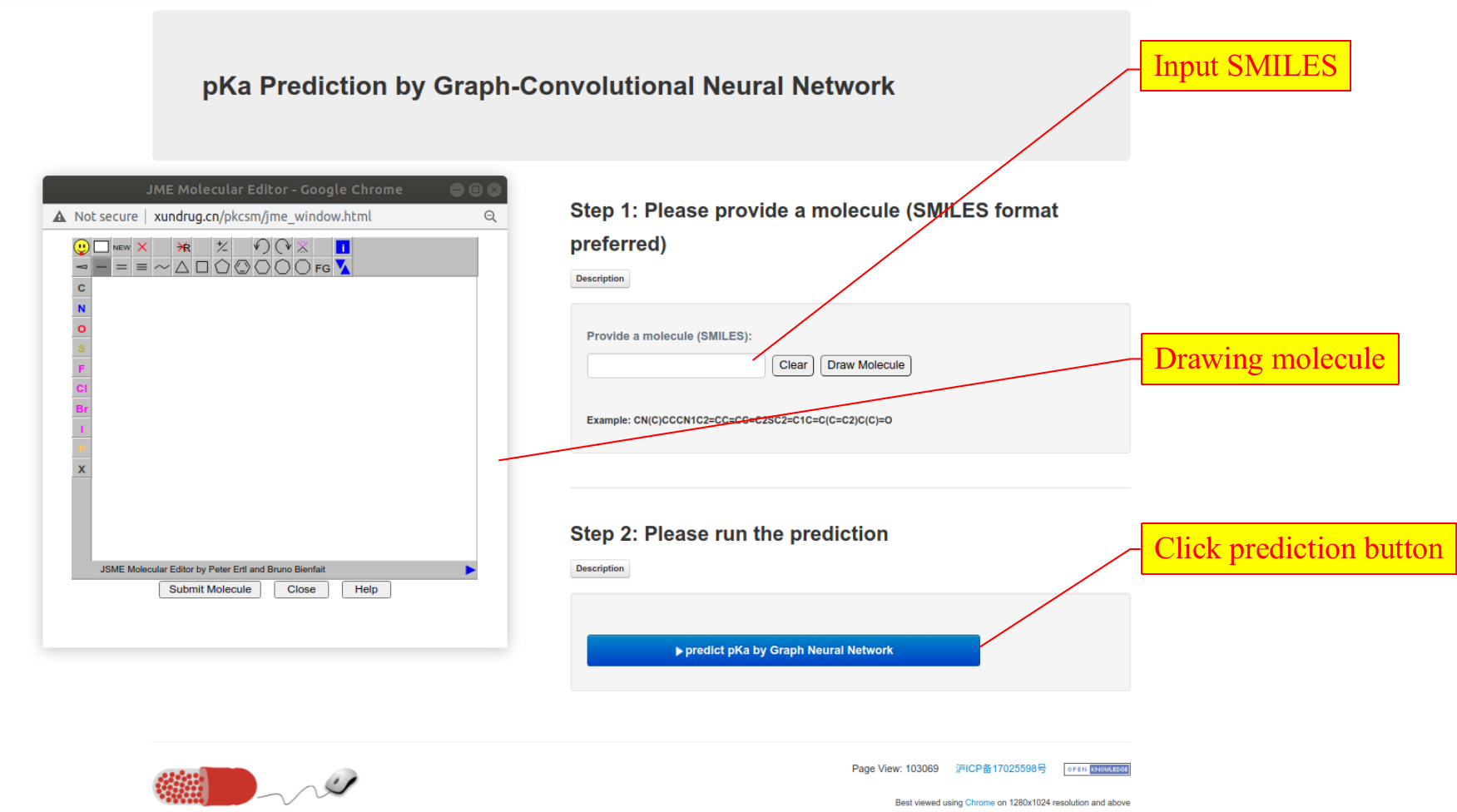
1. On the xundrug.cn/molgpka website, please input a molecule (SMILES format) and then click the predict button. You can input the SMILES string directly, or you can draw a molecule through the JME molecule editor.
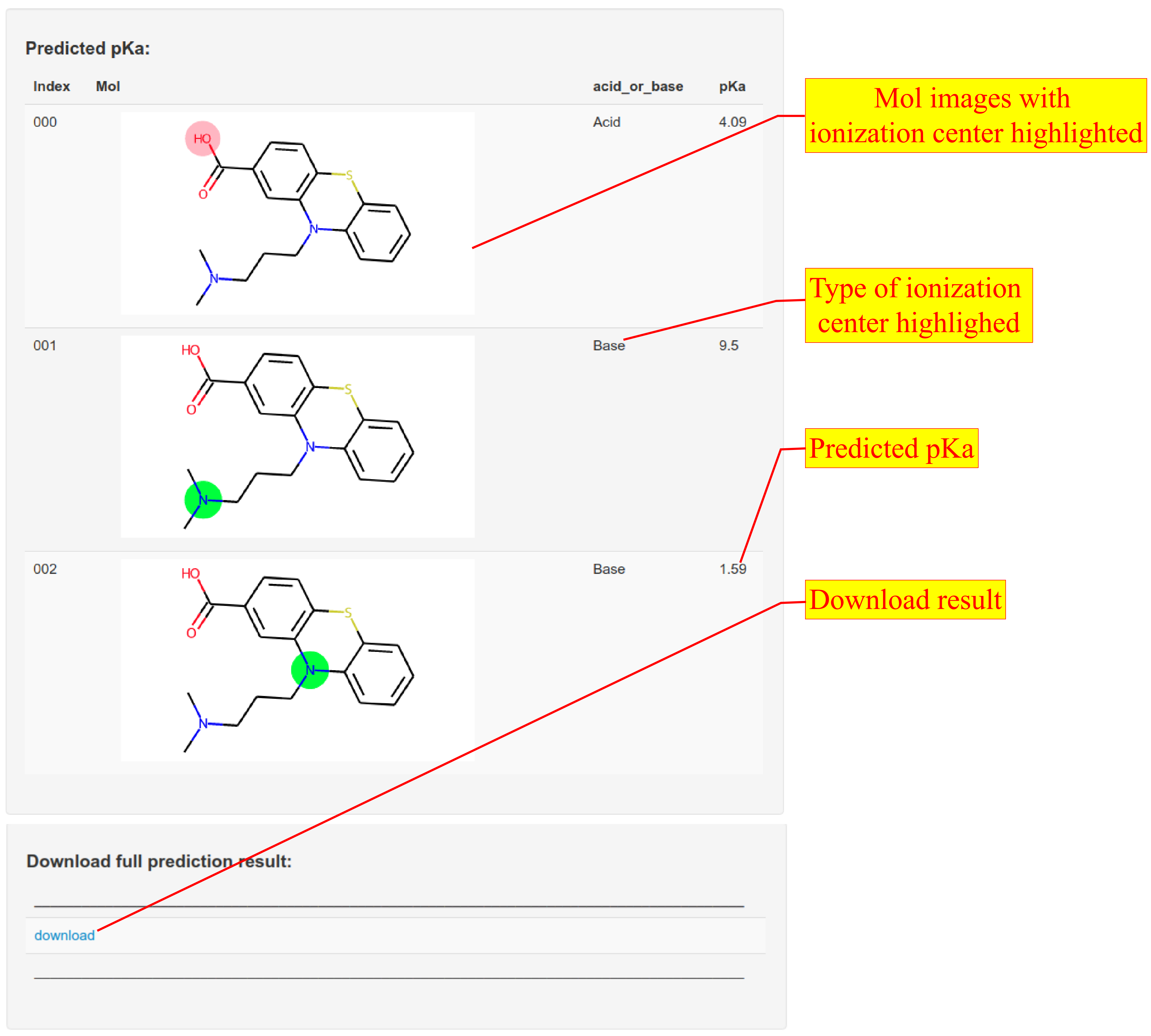
2. The server extracts all ionization centers on the input molecule and predicts pKa for each ionizable group by graph-convolutional neural network. The result page shows information of all ionization centers and predicted pka.
Example for pka prediction with MolGpKa API
To prevent overloading, please do not send over 10000 requests per day or 500 requests per hour.
Contact
If you have any question about MolGpKa, please contact:
Changge Ji
Chicago.ji@gmail.com